NVIDIA AI stack available now for researchers on Verily Pre
Verily has just announced additional apps and scalable hardware now available in Verily Workbench, including a suite of NVIDIA tools alongside enhanced support for the latest NVIDIA GPUs — NVIDIA DGX B200, NVIDIA H200 Tensor Core, and NVIDIA H100 Tensor Core GPUs. This development provides readily accessible GPU-powered libraries and tools, enabling researchers to accelerate analyses, undertake larger-scale projects, and more efficiently analyze Omics data and train Large Language Models (LLMs).
Verily Workbench, built on the Verily Pre platform, accelerates and elevates data analytics and governance for life science organizations and data providers. It achieves this through collaborative research features and infrastructure that streamline data analytics and governance across the enterprise, leading to quicker insights. A key feature of Workbench is providing users access to powerful, reproducible, and customizable computational analysis environments, known as cloud applications. These cloud apps come pre-configured with tools, such as JupyterLab, R Analysis Environment, and Visual Studio Code, eliminating the need for manual installations and setup.
Benefits of NVIDIA Parabricks, NVIDIA CUDA-X Data Science and NVIDIA NeMo on Workbench
Researchers can access two new cloud apps on Workbench: one app pre-configured with NVIDIA Parabricks and NVIDIA CUDA-X Data Science (CUDA-X DS), and a second cloud app pre-configured with NVIDIA NeMo Framework. NVIDIA Parabricks is a scalable genomics software suite for secondary analysis that provides GPU-accelerated versions of trusted, open-source tools. It provides GPU-accelerated genomic analysis tools for some of the most common analyses including read alignment and somatic and germline variant calling. NVIDIA CUDA-X DS is a collection of open-source libraries that accelerate popular data science libraries like pandas and scikit-learn. It can significantly accelerate research and data science workflows for tertiary analysis. Finally, NVIDIA NeMo Framework is a scalable and cloud-native generative AI framework built for researchers and PyTorch developers working on Large Language Models (LLMs), Multimodal Models (MMs), Automatic Speech Recognition (ASR), Text to Speech (TTS), and Computer Vision (CV) domains. It is designed to help you efficiently create, customize, and deploy new generative AI models by leveraging existing code and pre-trained model checkpoints.
Researchers can now easily deploy the two new NVIDIA-supported apps and interact with the libraries through JupyterLab
Quicker analysis — shorter turnarounds
NVIDIA Parabricks opens the door for new applications including the processing of sequencing data with much deeper coverage (e.g. 60x or 100x) for analysis of somatic or rare variants, and faster iteration and data processing of larger population-level cohorts and datasets.
Similarly, NVIDIA CUDA-X DS can offer tremendous value in the computational and machine learning stages of data manipulation and statistical modeling, including clustering and dimensionality reduction (e.g. PCA and UMAP). Scientists can leverage NVIDIA CUDA-X DS tools for genomic applications, such as Polygenic Risk Score (PRS) calculations, as well as Genome Wide Association Studies (GWAS and PheWAS).
NVIDIA NeMo completes this suite by providing a framework for generative AI and advanced natural language processing (NLP). It opens the door for users on Workbench to build and customize LLMs and multimodal models on their biomedical data at a much larger scale.
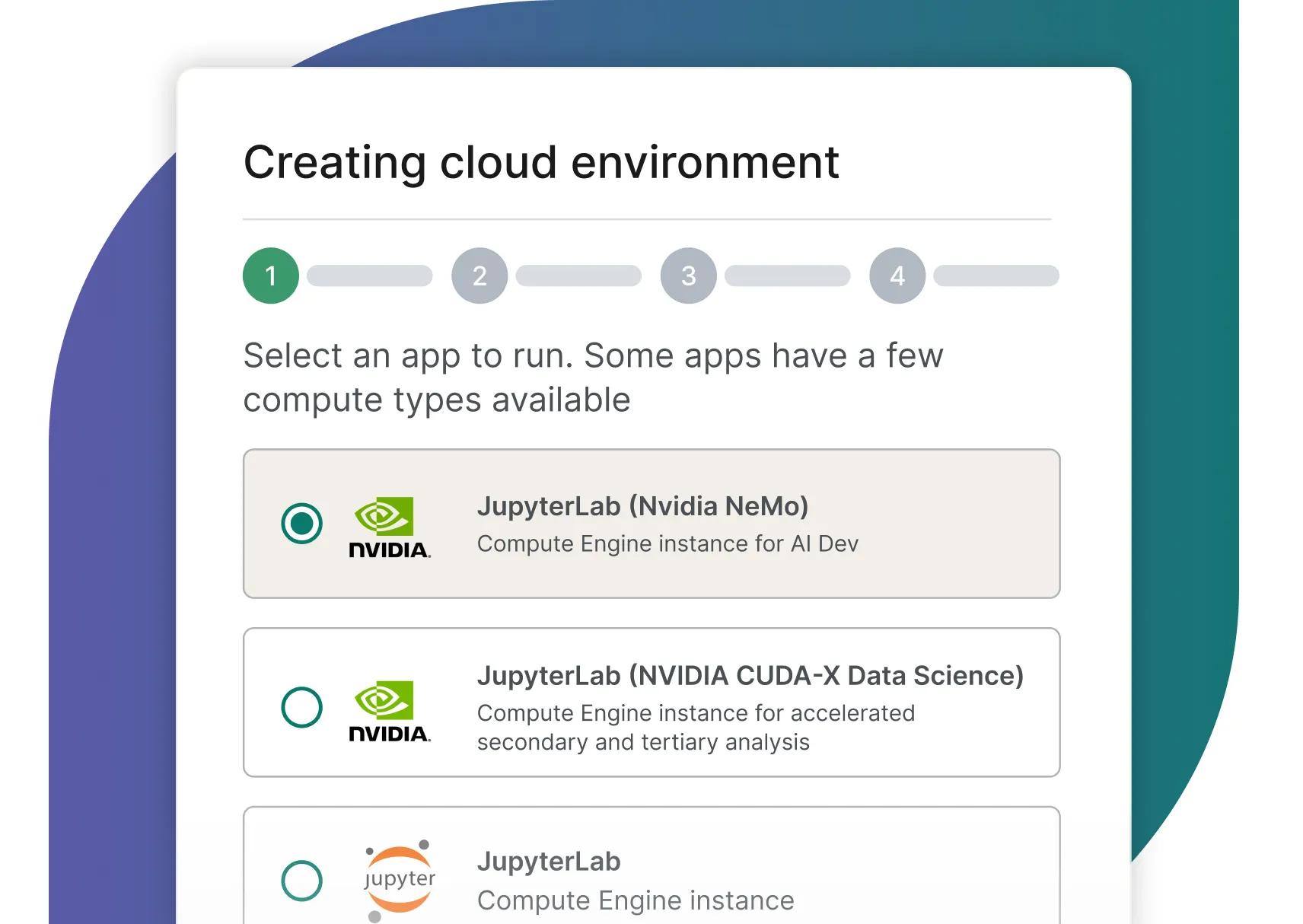
Convenient access to cloud apps pre-configured with NVIDIA NeMo, NVIDIA Parabricks and NVIDIA CUDA-X Data Science.
Unleashing performance with NVIDIA apps and modern GPUs
To maximize the value of the NVIDIA suite of tools, Verily added support for H100, H200, and Blackwell B200 GPUs. What does this mean? Users can now provision cloud apps, including the NVIDIA NeMo, NVIDIA Parabricks, and NVIDIA CUDA-X DS apps, with some of the most powerful GPUs offered on Google Cloud.
Note for users:
Given their popularity (e.g. H200, B200), users can make a specific or future reservation through Google Cloud. With a reservation name, users can easily consume it on Workbench – more documentation can be found here.
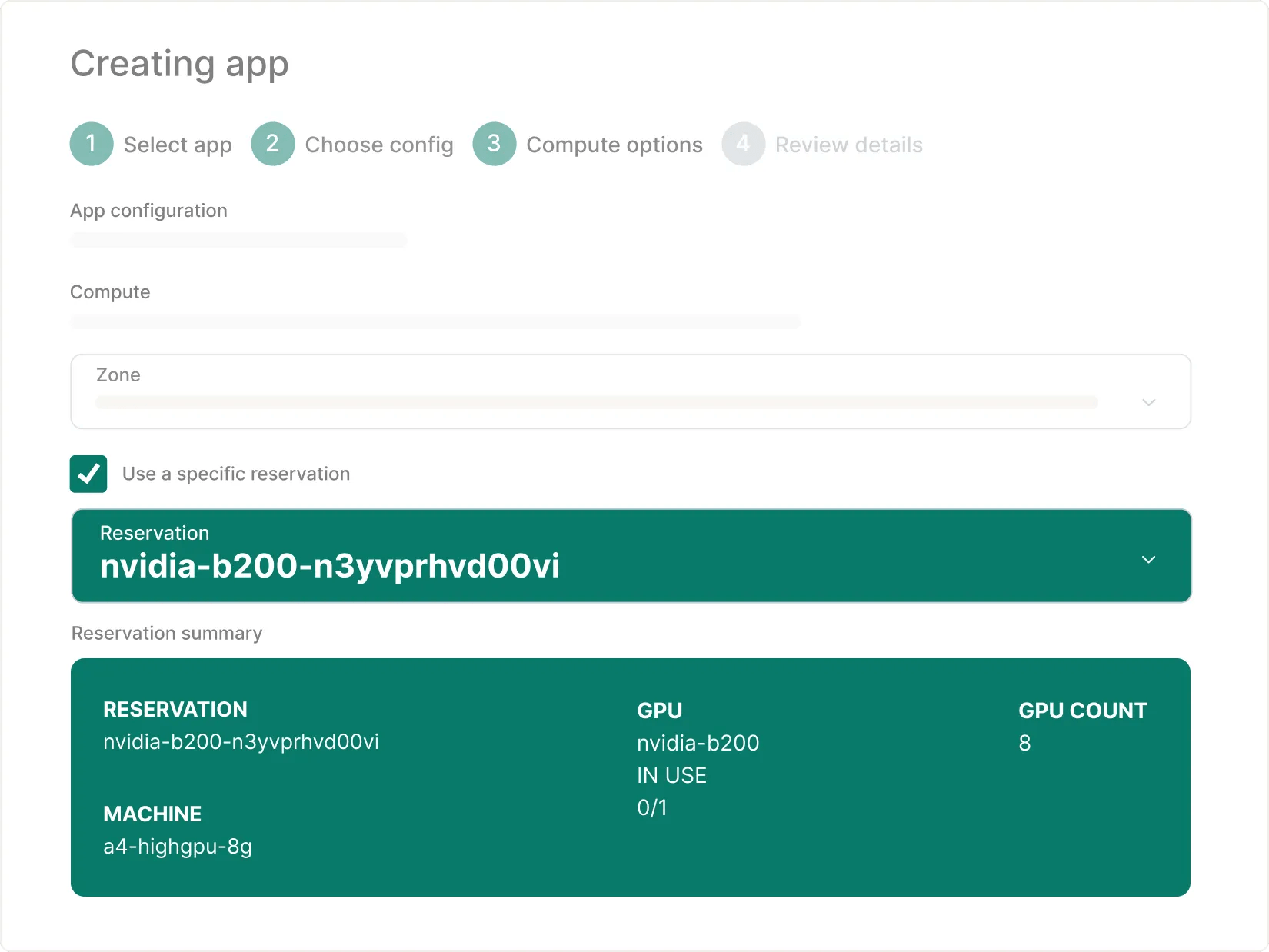
You can launch a cloud app with modern GPUs (e.g. B200) by simply referencing the reservation name.
Evaluating NVIDIA Parabricks with B200 Blackwell GPUs
Now that we have cloud apps pre-configured with NVIDIA libraries and access to cutting-edge GPUs, what kind of performance can our users expect? We ran tests using data from the NIH All of Us Research Program for an initial round of evaluations. The All of Us dataset is a uniquely large and multimodal longitudinal dataset from over half a million participants across the United States. The dataset is available on the All of Us Researcher Workbench for users that complete the registration and approval process.
We first created a workspace with access to the All of Us dataset. The test consisted of read alignment pipelines using GPU-accelerated and CPU-only cloud app configurations. The specific test parameters are as follows:
| Workflows | Sequencing Alignment | |
|---|---|---|
| Test Configuration | NVIDIA Parabricks fq2bam with B200 GPUs | BWA-MEM and GATK MarkDuplicates with 32 CPU threads |
| GCP Machine Type | a4-highgpu-8g | n2-standard-96 |
| Storage Disk | Locally mounted SSD | Locally mounted SSD |
| Results | 10m 2s | 369min 17s |
| Acceleration | 36.8X | 1X |
We tested the read alignment pipelines (FASTQ-to-BAM) using Whole-Genome Sequencing (WGS), generated with pair-end reads at 30X coverage from a sample belonging to the All of Us dataset. Specifically, we tested the Parabricks fq2bam pipeline with B200 GPUs (a4-highgpu-8g) against BWA-MEM and GATK (MarkDuplicates) on a CPU-only machine (n2-standard-96) running on 32 threads. To minimize any possible impacts due to storage disk I/O limits, both virtual machines (VMs) were mounted with SSD disks. All input files were copied onto the SSD disks. The runtime with NVIDIA Parabricks and the B200 GPUs was ~37X faster than BWA-MEM + GATK on the CPU 96-core machine.
Accelerating training for an All of Us foundation model using the pre-configured NVIDIA NeMo cloud app
The NVIDIA NeMo cloud app has been used to train a foundation model with the All of Us dataset found in this pre-print.
The team at Verily explored the overall performance of an electronic health record (EHR) foundational model (FM) with the integration of external modalities, namely genomic data through the use of Polygenic Risk Scores (PRS). The investigation included a performance evaluation of the model training steps themselves; using NVIDIA Nemo Automodel and H100 GPUs, Verily researchers were able to train the model 10x faster than previous approaches. In particular, the tests revealed an acceleration of ~3X when using NeMo Automodel vs. Hugging Face Accelerate on the same H100 GPU hardware – 5.0 steps/sec and 15.7 steps/sec, respectively.
Testing demonstrated the value of NVIDIA NeMo for FM training, and the benefits of accessing these pre-configured apps on Workbench in a secure and compliant manner (e.g. configured and approved for use with the All of Us dataset).
| Accelerate on H100 | NeMo Automodel on H100 |
|---|---|
| 5.0 steps/sec | 15.7 steps/sec |
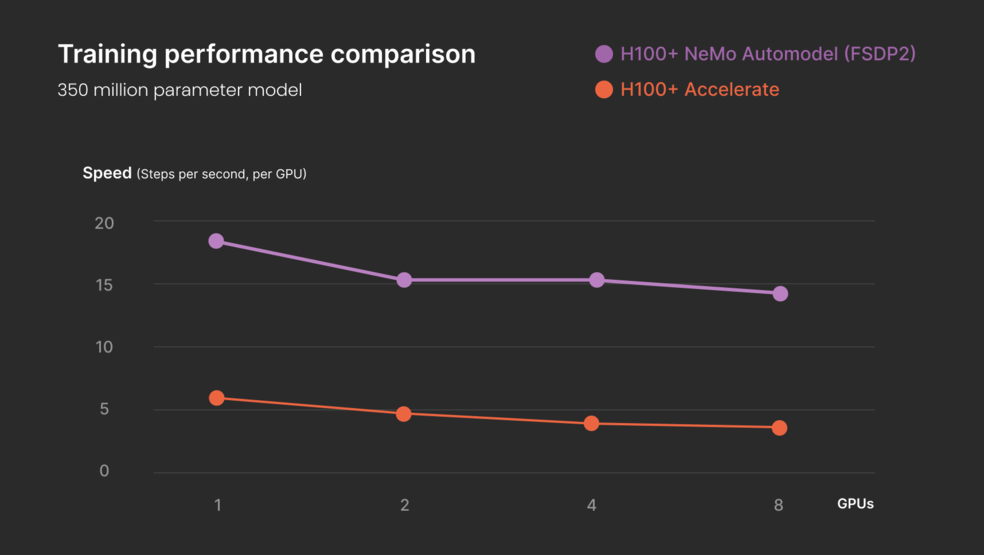
Performance comparison for the training of an EHR foundational model with the All of Us dataset
Configurations for the NVIDIA cloud apps on Workbench
The cloud apps pre-configured with NVIDIA NeMo and NVIDIA Parabricks and NVIDIA CUDA-X DS on Workbench use the devcontainers listed below:
Workbench users can also access our template to modify the apps as needed.
Preview: Running workflows on Workbench with NVIDIA Parabricks
One way to scale secondary and tertiary analyses leveraging the NVIDIA suite of tools is through the use of workflows: large-scale batch processing jobs that can apply thousands of machine-hours to massive computational jobs. Workbench offers a comprehensive set of features supporting workflow executions and monitoring capabilities.
Workflows allow you to describe your entire analysis as one single, repeatable, and shareable plan. Workbench provides support for workflow languages such as Workflow Definition Language (WDL), Nextflow and dsub in order to automate the execution of one or many steps of a data analysis pipeline.
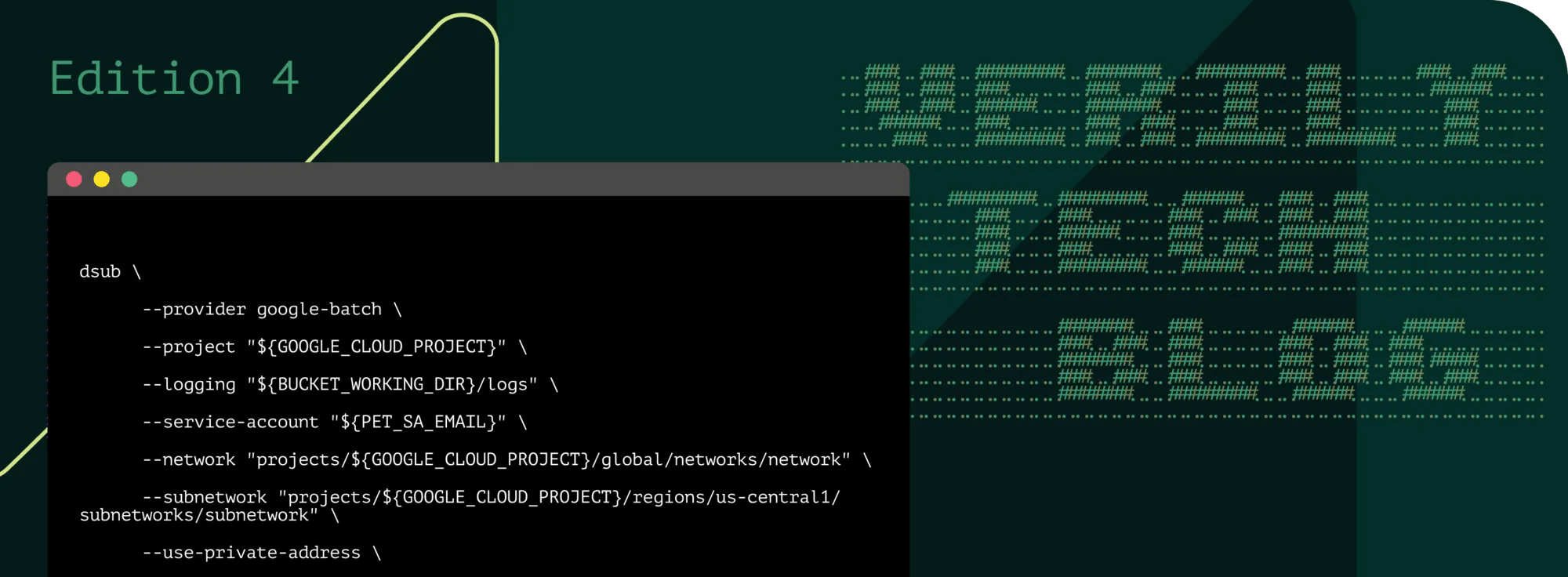
We successfully tested the execution of workflows with NVIDIA Parabricks and B200 GPUs through dsub using a beta release soon to be made publicly available. Briefly, dsub is an easy-to-use command-line tool designed to simplify running batch scripts particularly on Google Cloud, using Google Batch. Here’s an example command-line block to run these on Workbench with an A100 GPU:
This script submits a job to Google Batch that runs NVIDIA Parabricks fq2bam. It references a NVIDIA Parabricks image hosted in Google Artifact Registry, and runs on an a2-highgpu-1g machine with one NVIDIA A100 GPU. Input files are read from Google Cloud Storage buckets defined in the environment variables (REF_DIR, FASTQ_R1, FASTQ_R2).
There are many benefits that NVIDIA tools and frameworks like NVIDIA Parabricks, NVIDIA CUDA-X DS, and NVIDIA NeMo bring to Verily Workbench, alongside support for advanced GPUs such as the H100, H200, and B200. These additions significantly accelerate genomic analysis, data science workflows, and training of LLMs, demonstrating substantial performance gains. Ultimately, Workbench provides a secure and compliant environment for researchers. This enables users to access powerful NVIDIA capabilities and leverage vast, multi-modal datasets like the All of Us program, other generally available data from our exchange, or internal and proprietary data for our enterprise customers, driving new discoveries in life sciences R&D.